Let’s go over the basic structure of the portal so you can navigate it successfully. Landing on the portal, you’ll find a menu along the top with different tabs to choose from:
About, Data Access, Explore, News, and Help.

Let’s go through each one so you understand how to navigate the portal in a way that best suits your needs.
About
This is information about who funds and supports the Portal.
Data Access
Although anyone can view data readily from the portal (thanks to our open data sharing model), you need a Synapse account in order to download data.
Data held in the portal falls into two categories: Open and Controlled Use. While Open data is available for all registered Synapse users without limitations, Controlled Use data is available to registered, certified, or validated Synapse users that fulfill specific requirements for data access, including a signed Data Use Certificate and Intended Data Use Statement.
Find instructions to register for a Synapse account and to submit a DUC on the the Data Access page
Explore
The Explore tab isn’t a page on its own, but a menu of subtabs/pages to choose from:

These items correspond to the different ways in which you can find information about and filter and view data.
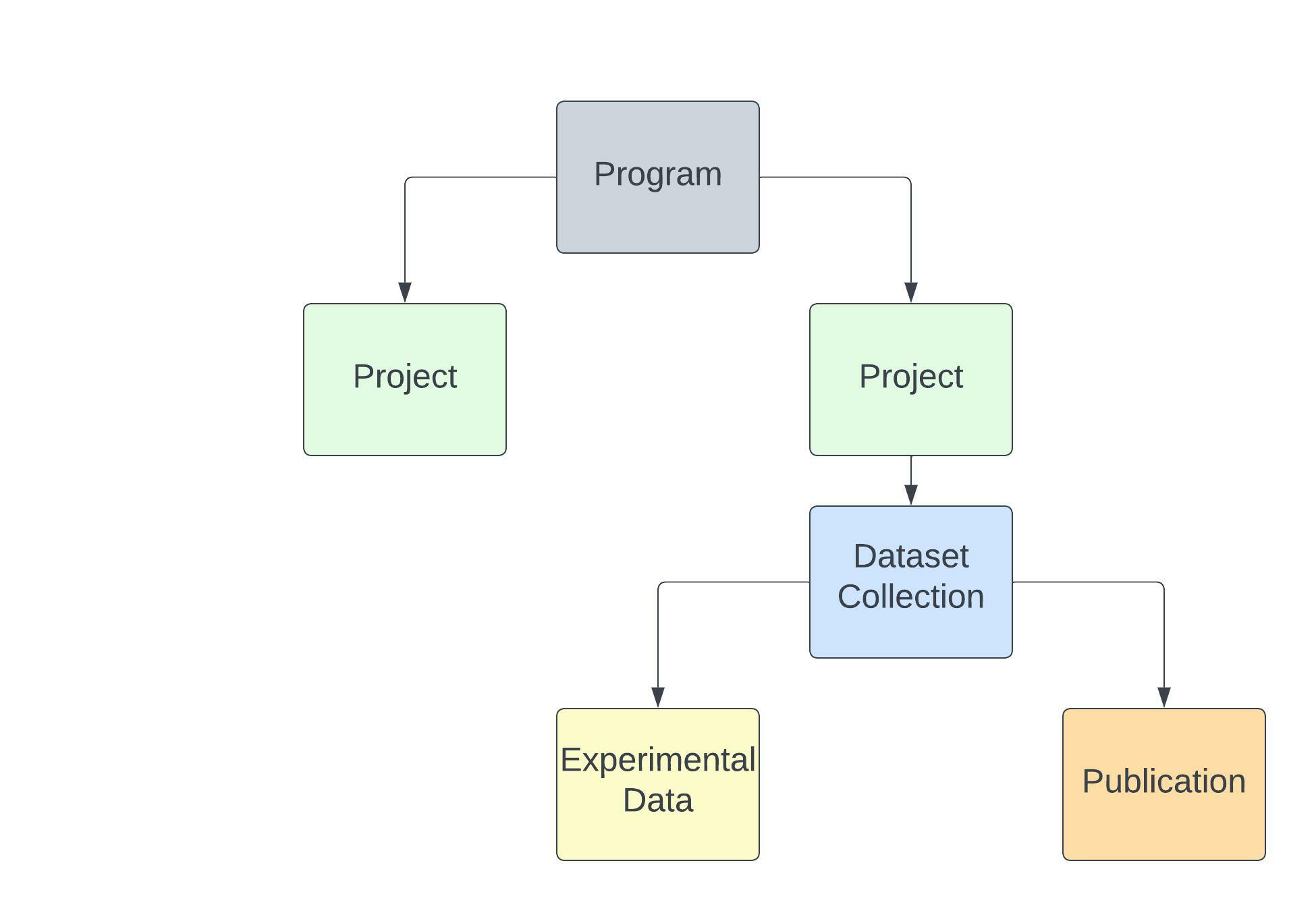
For example, the Programs page lists all research programs, so you can read about and select one of interest. A Program may have one or multiple Projects that describe further details of the Program. Data in the Portal are grouped into Datasets, which are linked back to the Project they were generated through. There are 2 types of Datasets (Collections). Experimental Data - which is are files related to specific assays, and/or diseases, and Publications - which are files used in specific manuscripts. The All Data tab allows for exploration of all files in the portal irrespective of which Program, Project, or Dataset the files are associated with.
News
The News tab links to a page providing data release notes.
Help
This tab brings you to this help site!